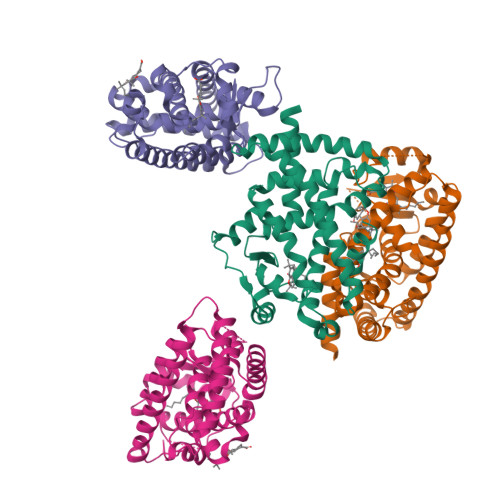
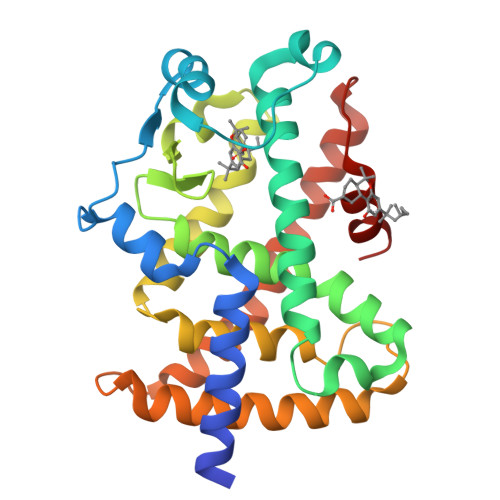
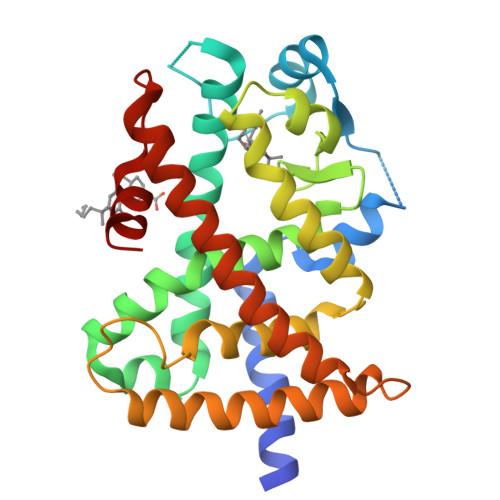
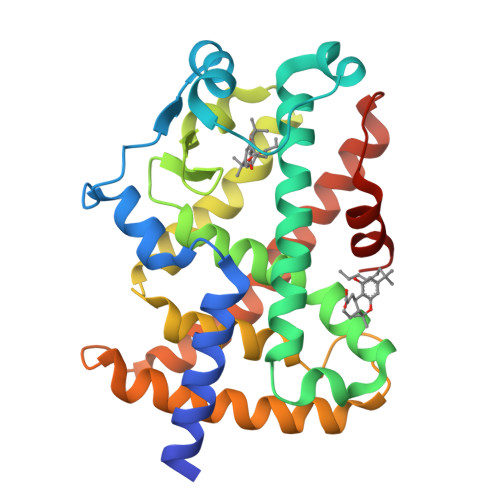
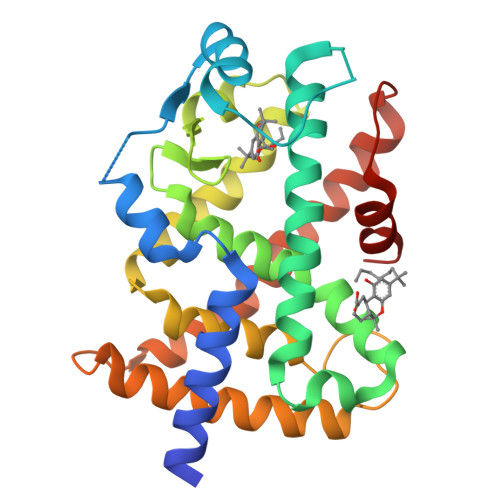
Ajulemic Acid, a Synthetic Nonpsychoactive Cannabinoid Acid, Bound to the Ligand Binding Domain of the Human Peroxisome Proliferator-activated Receptor gamma
Ambrosio, A.L.B., Dias, S.M.G., Polikarpov, I., Zurier, R.B., Burstein, S.H., Garratt, R.C.(2007) J Biological Chem 282: 18625-18633
- PubMed: 17462987
- DOI: https://doi.org/10.1074/jbc.M702538200
- Primary Citation of Related Structures:
2OM9 - PubMed Abstract:
Ajulemic acid (AJA) is a synthetic analog of THC-11-oic acid, a metabolite of tetrahydrocannabinol (THC), the major active ingredient of the recreational drug marijuana derived from the plant Cannabis sativa. AJA has potent analgesic and anti-inflammatory activity in vivo, but without the psychotropic action of THC. However, its precise mechanism of action remains unknown. Biochemical studies indicate that AJA binds directly and selectively to the isotype gamma of the peroxisome proliferator-activated receptor (PPARgamma) suggesting that this may be a pharmacologically relevant receptor for this compound and a potential target for drug development in the treatment of pain and inflammation. Here, we report the crystal structure of the ligand binding domain of the gamma isotype of human PPAR in complex with ajulemic acid, determined at 2.8-A resolution. Our results show a binding mode that is compatible with other known partial agonists of PPAR, explaining their moderate activation of the receptor, as well as the structural basis for isotype selectivity, as observed previously in vitro. The structure also provides clues to the understanding of partial agonism itself, suggesting a rational approach to the design of molecules capable of activating the receptor at levels that avoid undesirable side effects.
Organizational Affiliation:
Centro de Biotecnologia Molecular Estrutural, Instituto de Física de São Carlos, Universidade de São Paulo, São Carlos-SP CEP 13560-970, Brazil. Electronic address: ala48@cornell.edu.